Research Interests †
Thermococcus kodakarensis KOD1 †
Thermococcus kodakarensis KOD1 is a hyperthermophilic archaeon isolated from a geothermal spring near the coast of Kodakara Island, Kagoshima, Japan by the group of Prof. Tadayuki Imanaka. It has previously been known as Pyrococcus sp. KOD1, Pyrococcus kodakaraensis KOD1, and Thermococcus kodakaraensis KOD1. The organism grows at temperatures between 60 and 100˚C, with an optimum growth temperature of 85˚C. It is an obligate heterotroph, and can utilize a variety of organic compounds such as amino acids, peptides, maltooligosaccharides, starch and organic acids such as pyruvic acid. When grown on amino acids, elemental sulfur is necessary as an electron acceptor, resulting in the generation of hydrogen sulfide. When the organism is grown on sugars or pyruvic acid, electrons are given to protons, generating molecular hydrogen.
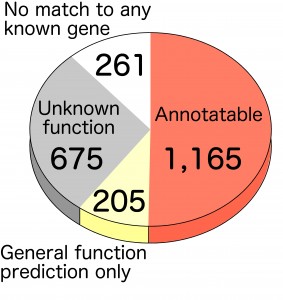
We have determined the entire genome sequence of T. kodakarensis. It consists of 2,088,737 bp and harbors a predicted 2,306 open reading frames. The function of roughly half of these genes can be predicted by primary structure, while the functions of the other half are unknown. Our primary goals are to determine the physiological roles of genes whose functions are unknown, with emphasis on metabolism and transcriptional regulation. We have also experienced in many cases that functional prediction based on sequence data is not necessarily accurate, and when so, we try to correct these predictions based on experimental evidence. In addition to sequencing the genome, we have developed a genetic system based on homologous recombination in T. kodakarensis. This allows the disruption of any given non-essential gene, and can also be utilized for other genetic manipulations such as gene insertion and promoter exchange. The genome information and genetic system in T. kodakarensis has enabled us to study the function of many genes both in vitro and in vivo.
In addition to physiology studies on T. kodakarensis, we are studying a number of other organisms, which include halophiles, methanogens, hyperthermophilic bacteria and uncultivated organisms whose genome data are available through metagenome analyses. We are also actively engaged in exploring new possibilities for the application of hyperthermophiles and their enzymes in biotechnology. Examples would be developing in vitro transcription/translation systems using the machinery in (hyper)thermophiles and microbial hydrogen production from biomass.




![[logo] [logo]](wiki/image/logo.jpg)
